Challenge
Quadratic Knapsack Problem
Gene cluster analysis: Understanding genetic diseases.
Performance Overview
Speed vs IHEA
85×
faster execution
Speed vs QKBP
4.1×
faster execution
Quality vs QKBP
21×
better solution quality
This chart shows the solution quality evolution using the Relative Percentage Deviation (RPD) from optimal solutions, displayed on a logarithmic scale. Lower RPD values indicate better performance, with our transformation positioning better quality higher on the plot. Results are combined from testing on benchmark instances ranging from size 100 to 10,000 (StandardQKP, QKPGroupII, QKPGroupIII, and LargeQKP).
From round 64 onwards, TIG algorithms demonstrate superior solution quality compared to established state-of-the-art methods. The consistent improvement in RPD values shows how our competitive ecosystem drives algorithmic innovation, achieving solutions that are 21× better than QKBP (2025).
The dramatic improvement from round 44 to 64 illustrates the power of iterative refinement within the TIG protocol, where each algorithm builds upon previous innovations while competing for rewards. Read the complete research analysis for detailed methodology and benchmarking results.
Problem Overview
The quadratic knapsack problem is one of the most popular variants of the single knapsack problem, with applications in many optimization contexts.
The aim is to maximize the value of individual items placed in the knapsack while satisfying a weight constraint. However, pairs of items also have positive interaction values, contributing to the total value within the knapsack.
Challenge Overview
For our challenge, we use a version of the quadratic knapsack problem. The size of the challenge instance is determined by the parameter , which is the number of items from which you need to select a subset to put in the knapsack.
The weight of each of the is an integer, chosen independently, uniformly at random, and such that each of the item weights , for . The values of the items are nonzero with a density of 25%, meaning they have a 25% probability of being nonzero. The nonzero individual values of the item, , and the nonzero interaction values of pairs of items, , are selected at random from the range .
The total value of a knapsack is determined by summing up the individual values of items in the knapsack, as well as the interaction values of every pair of items , where , in the knapsack:
We impose a weight constraint , where the knapsack can hold at most half the total weight of all items.
The quality of a solution is determined by how much better it is than a baseline value. The baseline value is determined by using a baseline algorithm. In particular a solution is given a quality score defined as . For precision, `better_than_baseline` is stored as an integer where each unit represents 0.01%. For example, a value of 150 corresponds to 150/10000 = 1.5% improvement over the baseline value.
Example
Consider an example of a challenge instance with . Let the baseline value be 46:
weights = [39, 29, 15, 43]
individual_values = [0, 14, 0, 75]
interaction_values = [ 0, 0, 0, 0
0, 0, 32, 0
0, 32, 0, 0
0, 0, 0, 0]
max_weight = 63The objective is to find a high value set of items where the total weight is at most 63. Now consider the following selection:
selected_items = [2, 3]When evaluating this selection, we can confirm that the total weight is less than 63, and the total value is 75,
- Total weight = 15 + 43 = 58
- Total value = 0 + 75 + 0 = 75
Applications
The Knapsack problems have a wide variety of practical applications. The use of knapsack in integer programming led to breakthroughs in several disciplines, including energy management and cellular network frequency planning.
Although originally studied in the context of logistics, Knapsack problems appear regularly in diverse areas of science and technology. For example, in gene expression data, there are usually thousands of genes, but only a subset of them are informative for a specific problem. The Knapsack Problem can be used to select a subset of genes (items) that maximizes the total information (value) without exceeding the limit of the number of genes that can be included in the analysis (weight limit).
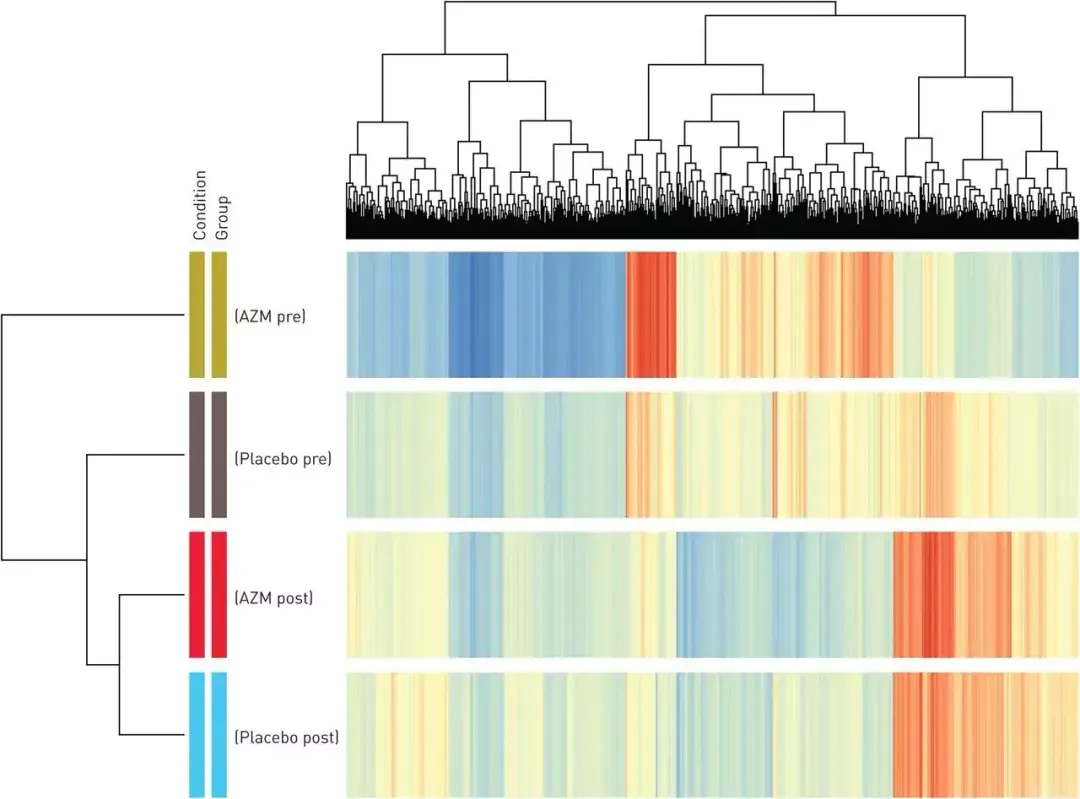
Figure 1: Microarray clustering of differentially expressed genes in blood. Genes are clustered in rows, with red indicating high expression, yellow intermediate expression and blue low expression. The Knapsack problem is used to analyse gene expression clustering.